I heard that RNA methylation is very hot recently. Is it sacred? Let us take a look at the specific project in 2017. We can also see that in addition to identifying the entry point of RNA methylation-related enzymes, the topic of RNA methylation covers a wide range of topics, including: disease development, rice reproductive development. , fruit fly sex determination and many other important areas. Since the field of RNA methylation is very new, the profile of RNA methylation in most species, tissues and diseases has not been reported. For those who want a fast track teacher who wrote, RNA methylation profiling is very appropriate. The article on the m5C RNA-modified expression profile is here, and it can be seen that almost no experimental manipulation is involved. It is only a detailed demonstration and analysis of the results of m5C RNA methylation sequencing. The highlight is that combined with CLIPdp data predicts the potential of the m5C RNA methylation site as an RBP target. It is worth learning from. Figure: Knockdown of ALKBH5 upregulates FOXM1 precursor mRNA expression and m6A RNA modification Figure: RIP and RNA Pull down experiments demonstrate direct binding of ALKBH5 and FOXM1-AS Finally, the authors further studied the effect of FOXM1-AS on the biological function of malignant glioma cells. The results showed that FOXM1-AS can positively regulate the expression of FOXM1 and maintain the performance of malignant glioma cells. Exogenous FOXM1 significantly reversed the inhibition caused by knockdown of ALKBH5.
Cannulated screw is a common internal fixation device in orthopedic surgery.
Working process
During the operation, the Kirschner wire with the hole matching the hollow screw is first drilled into the bone of the fracture site, and then the hollow screw is screwed into the fracture site through the guidance of the Kirschner wire. Finally, the Kirschner wire is pulled out to complete the fixation of the fracture site. In many cases of fracture of the neck and femur, the healing cycle is long, and it usually takes about 30 months to remove the cannulated screws. At this point, the bone and the surface of the cannulated nail are closely connected and firmly connected in the nail path, which leads to defects easily generated by conventional nail removal methods, causing secondary injury at the fracture healing site and causing pain to the patient. Percutaneous cannulated screw internal fixation for femoral neck fractures has the advantages of small trauma, good healing, low rate of femoral head necrosis, and good stability.
compression fracture,Cannulated Screws,surgery screws,Orthopedic Screw Jiangsu Aomed Ortho Medical Technology Co.,Ltd , https://www.aomedortho.com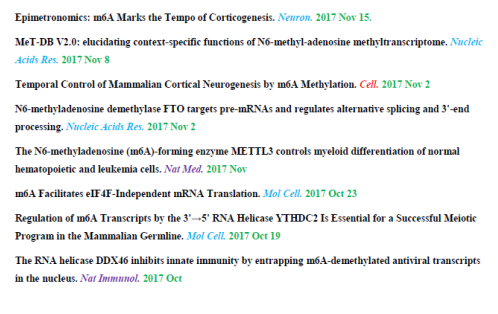

Scope of application: m5C RNA methylation; 

2018 National Nature Research Hotspot 2: In-depth analysis of RNA methylation research
1, high scores frequently appear
Speaking of recent research hotspots, the research on RNA methylation modification can be said to be one of the most popular directions in the whole life science field. The highlights of the articles are frequent, and it is really dizzying. Research on RNA methylation In the past three months , the article had an impact factor of 10 or more, and there were as many as 17 articles.
Figure: A recent look at the recent high-scoring article on RNA methylation
2, the number of articles has repeatedly climbed new heights
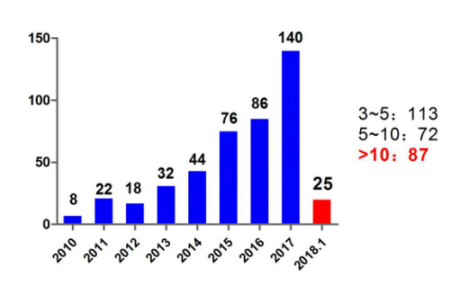
An increasing number of published articles, especially high-scoring articles, indicate that this field is quickly becoming the focus of attention. As for the publications over the years, the growth momentum in the field of RNA methylation is also very gratifying. In January 2018, only 25 published articles have been published.
Figure: RNA methylation over the years to publish article data
3, the country's natural project number rises gratifying
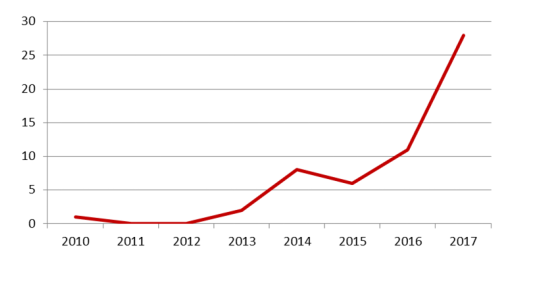
Vertically comparing the number of projects over the years, we can find that the fund project of RNA methylation has shown an exponential growth trend in the past two years: especially from 5 in 2015 to 25 in 2017, the project growth is particularly significant.
Figure: RNA methylation over the years
As can be seen from the above figure, the total number of projects related to RNA methylation research in the National Natural Science Foundation approved in 2017 is 25. From the research contents of the approved projects, there are some problems in the identification of RNA methylation profiles, the topic of RNA methylation modification enzymes, and the development of functional mechanisms for RNA methylation. This suggests that the study of RNA methylation is still in a state of basic screening and functional mechanisms that are relatively diverse. It is worth mentioning that there are also topics linking RNA methylation modification to miRNA and exocrine secretion to screen/identify tumor molecular markers, which is also an important direction for RNA methylation.
Figure: RNA methylation modification 2017 natural project enumeration
Second, I heard that there are three kinds of RNA methylation modification hot, who are they?
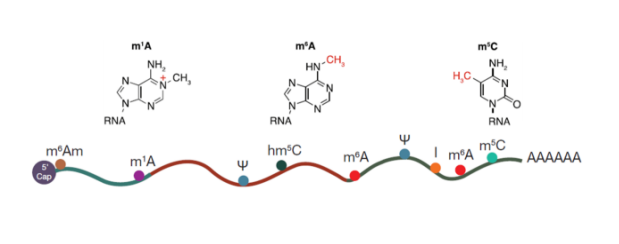
There are many types of RNA methylation modifications, and the three most popular ones are m6A RNA methylation, m5C RNA methylation, and m1A RNA methylation.
Figure: 3 popular RNA methylation modifications
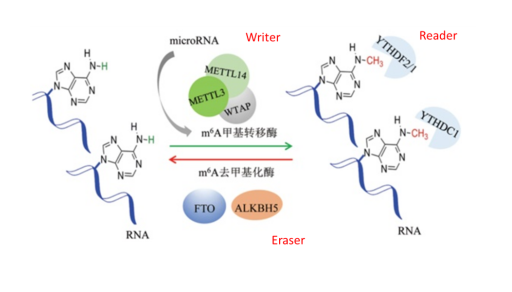
1. m6A RNA methylation is the most common and abundant post-transcriptional modification of eukaryotic mRNA. The current research is mainly focused on the key enzymes of each modification. Prof. He Chuan summarized the related modifications of m6A in the January 2017 Nature Review (IF=46.602) , consisting of methyltransferase complexes (METTL3, METTL14 and WTAP), demethylases (FTO and ALKBH5), and The corresponding readers (YTHDF1/2/3, YTHDC1) are coordinated. Studies of related functions have demonstrated that m6A modifications are dynamically reversible and have a wide variety of important biological implications. M6A RNA methylation has been found to have many functions, such as it can promote circular RNA translation (2017.3 Cell Res, IF 14.812) , or regulate cancer stem cell differentiation by promoting mRNA degradation (2016.4 Cell Host Microbe, IF9.661) It also regulates T cell differentiation and immune homeostasis (2017.8 Nature. IF 40.137) .
Figure: m6A RNA methylation dynamic reversible modification
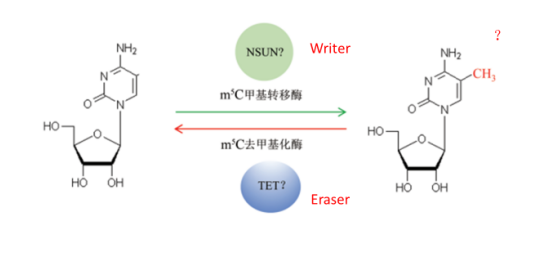
2. M5C RNA methylation is a recent discovery of a class of methylation modifications in the high abundance of tRNA and rRNA. High-throughput sequencing was used to verify the widespread presence of non-coding RNA and some mRNAs in m5C, but there is no systematic report on the distribution of m5C in different species and tissues. For m5C, there are also a series of related modified enzymes (as shown below). The m5C RNA methylation function has been found to regulate mRNA nucleation (2017 Cell Res, IF 14.812) or to regulate stem cell protein synthesis (2017 Nature, IF 40.137) .
Figure: m5C RNA methylation dynamic reversible modification
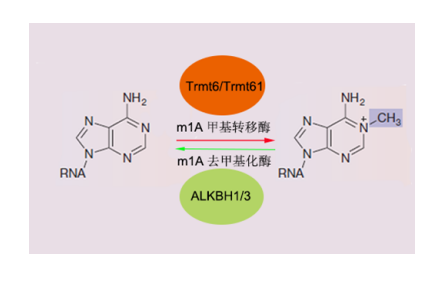
3. m1A RNA methylation is a new type of RNA methylation modification that enters everyone's field of view. A recent article on m1A RNA methylation published on Molecular Cell (IF: 14.714) describes a novel class of RNA modifications that are ubiquitous in non-coding RNAs and mRNAs. m1A RNA methylation function, it has been found that a variety of m1A modifications occur on tRNA (2017.2 Nucleic Acids Research, IF 9.28) , or participate in the regulation of translation initiation and extension (2016 Cell, IF 30.41) . In conclusion, RNA methylation is also an important way to regulate gene expression.
Figure: m1A RNA methylation dynamic reversible modification
3. What are the enzymes related to RNA methylation?
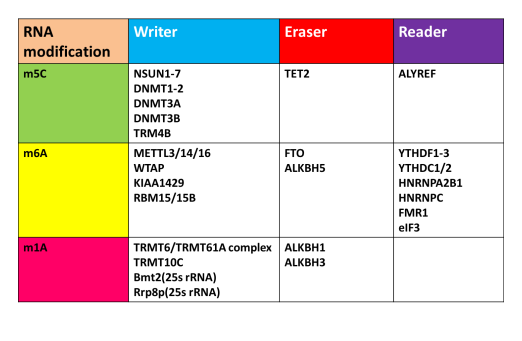
RNA methylation-related proteins contain a specific methylation-specific methyltransferase (Writer), a demethylase (Reader), and a corresponding RNA methylation recognition protein (Reader) for coordinated regulation to maintain a specific type A Dynamic modification of the base. Many high-scoring articles are aimed at methylation-modified proteins for functional mechanism studies. So what are the key modified proteins for different methylation modifications so far? After looking through the literature search, Xiaobian helped you summarize the relevant modified proteins reported in the current literature, which is convenient for everyone to refer to:
Figure: RNA methylation modification related enzyme finishing
Fourth, RNA methylation is so hot, what is its research idea?
The current research ideas of RNA methylation are shallow and deep, and there are two major categories: 1. RNA methylation profile 2. Functional mechanism research. Then let us divide them into different categories and learn!
1, RNA methylation spectrum
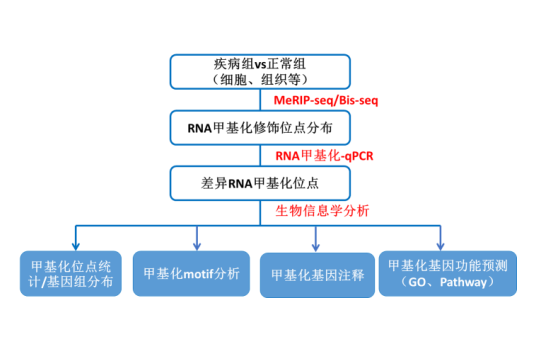
How is an article on RNA methylation spectrum refined? At the beginning of the study, we need to determine the research objectives: species, disease model, sample type, and as much detailed sample information as possible. Next, high-throughput screening of RNA methylation regions or loci by RNA methylation sequencing and in-depth bioinformatics mining, such as methylation site statistics, genomic distribution, and methylation function prediction (GO, Pathway analysis) and so on.
Figure: RNA methylation expression profile
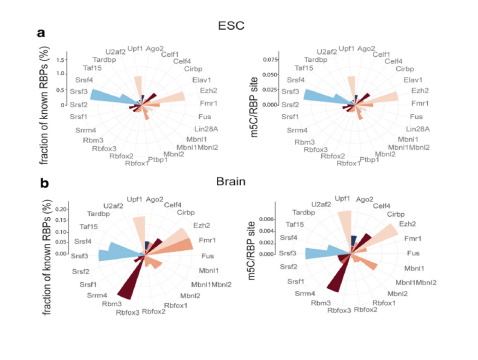
Let's take a look at a typical RNA methylation profile with an impact factor of 11 for example. This article, published in Genome Biology , shows the overall status of m5C RNA modification in mouse embryonic stem cells and brain.
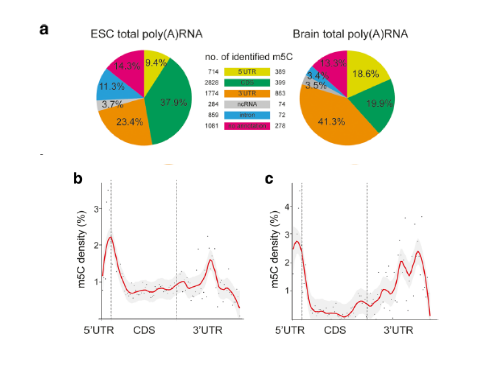
Figure: m5C RNA methylation site statistics
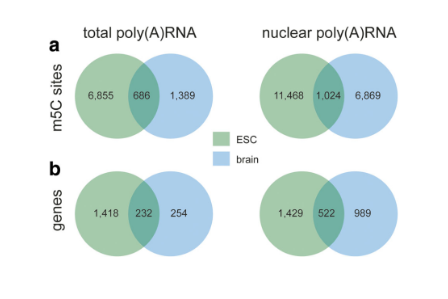
Co-analysis and specificity analysis of m5C loci of total poly(A) RNA and nuclear poly(A) RNA in different samples were first performed by Bis-seq technique.
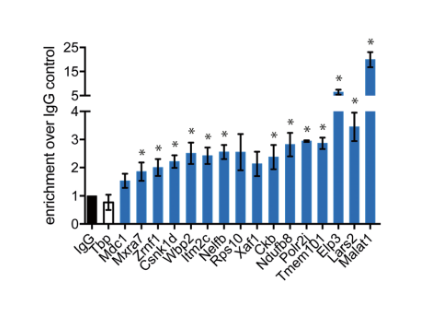
Figure: qPCR verification of m5C RNA methylation sequencing results
The m5C modification site was subsequently verified by qPCR, and it can be seen from the figure that 13 of the 16 verified transcripts were significantly enriched. Next, the results of m5C RNA methylation sequencing were analyzed. It was found that the specific m5C methylation modification in most ESC total poly(A) RNA was caused by the difference in methylation degree, and the genetic difference between different samples. Expression is irrelevant. Functional prediction of the corresponding gene of m5C-modified RNA revealed the top ten GO entries shared and unique between mouse brain tissue and mouse embryonic stem cells.
Figure: Distribution of m5C loci in different samples
Further data analysis showed that the distribution of m5C sites in the total poly(A) RNA of different samples was consistent: the m5C modification site in mouse embryonic stem cells was preferentially distributed near the mRNA start codon, and in mouse brain tissue. It tends to be distributed in the 3'UTR area.
Figure: CLIPdb: Co-analysis of m5C locus and RBPs binding sites
A combined analysis with CLIPdb revealed that approximately 29% of the poly(A)m5C RNA modification sites in mouse embryonic stem cells coincide with the corresponding RBP (RNA binding protein) binding sites, which is approximately in mouse brain tissue. At 11%, it shows the potential to bind to RBP and exert biological functions in specific transcripts.
2. Functional mechanism research
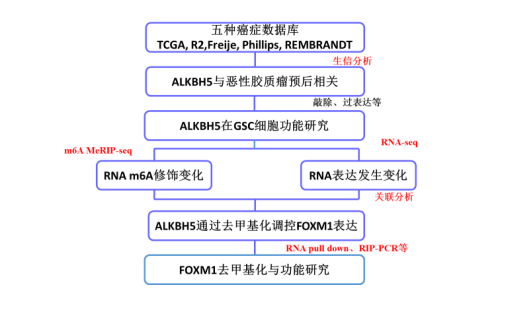
After reading the expression profile, how should the functional mechanism of RNA methylation be studied? Let's take a look at a Cancer Cell article with an influence factor of 27. Let's take a look. This is a collaboration between the Huang Suyun team from the Anderson Cancer Center in the United States and the Hechuan team at the University of Chicago, revealing the role of m6A RNA methylation in gliomas.
First put a technical roadmap.
Figure: Showcased article feature set technology roadmap
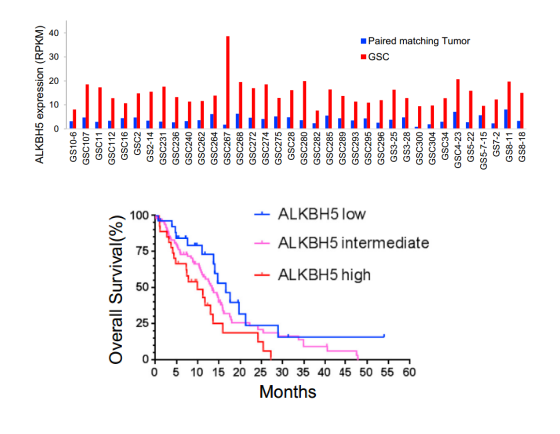
The TCGA database showed that the expression level of m6A demethylase ALKBH5 in GSC was significantly increased. For prognosis testing of different patients, the survival curve found that patients with high expression of ALKBH5 had a worse prognosis.
Figure: ALKBH5 is associated with poor clinical outcomes
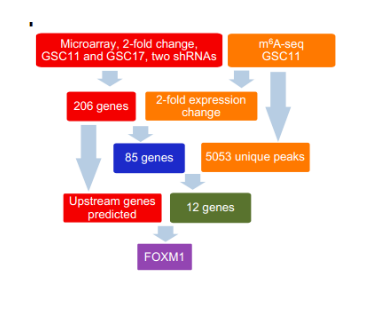
The authors identified the m6A RNA methylation modification map by MeRIP-Seq technology and combined the expression profiles to detect genes with >2 fold differential expression. The ALKBH5 target gene-FOXM1, which is involved in cell cycle regulation, was finally screened, which plays a key role in self-repair and tumor formation of GSCs.
Figure: ALKBH5 target gene - FOXM1 screening flow chart
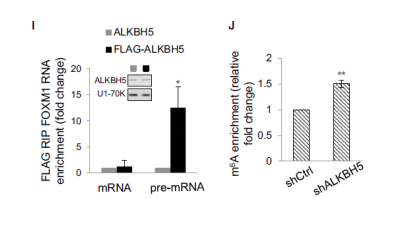
Knockdown of ALKBH5 by MeRIP qPCR increased the m6A methylation level of FOXM1 precursor mRNA. These results indicate that ALKBH5 primarily affects the expression of FOXM1 by demethylation activity.
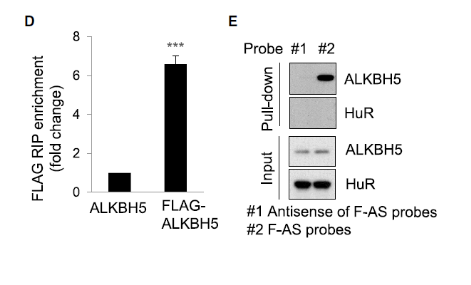
The RIP and RNA pull down experiments indicated that FOXM1-AS can directly bind to ALKBH5 and FOXM1 precursor mRNA.
V. Common techniques for RNA methylation research
1, cloud sequence organism MeRIP-Seq
Scope of application: m6A RNA methylation, m5C RNA methylation, m1A RNA methylation;
Technical principle: High-throughput sequencing is performed after enriching methylated RNA with specific antibodies;
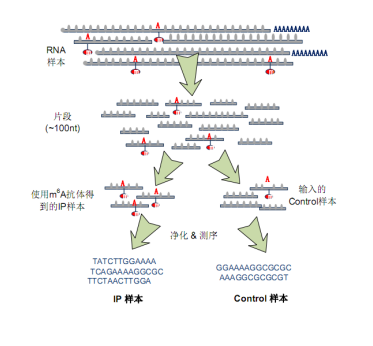
The most popular detection tool for RNA methylation is the MeRIP-Seq technology, which combines methylation RNA co-precipitation and RNA sequencing techniques to detect RNA methylation modifications in the entire transcriptome with high precision. The specific principle is as follows: an antibody against methylation of a specific RNA is co-incubated with an RNA sample (IP), and a methylated RNA fragment is captured for sequencing; and a parallel (Input) sample is required to be sequenced for elimination. Non-specific capture of the background of the methylated fragment. Comparing the sequence fragments in the immunoprecipitation sample (IP) and the control sample (Input), the RNA methylation modification region was mapped to the transcriptome, and the degree of RNA methylation in the sample was calculated. The cloud sequence organism has a comprehensive m6A, m5C, m1A methylation MeRIP-Seq product line, which can comprehensively detect various types of RNA methylation in circRNA, LncRNA and mRNA.
Figure: Cloud Sequence Bio MeRIP-seq Experimental Process
2, cloud sequence biology Bis-Seq
Technical principle: High-throughput sequencing detection of heavy sulfite-transformed RNA;
Modifications for m5C RNA methylation also have a single base resolution assay: samples are treated with bisulfite. Bisulfite treatment can convert C-bases that have not been methylated in the genome into U, and undergo PCR amplification to become T, so that the products of reverse transcription will undergo single-point mutation, combined with high-throughput sequencing technology. By performing sequence alignment with the untreated group, it is easy to find the m5C modified site. Cloud sequence organism has mature Bis-seq experimental platform and rich experience in RNA methylation sequencing, which can meet customers' needs for different samples and different types of RNA methylation sequencing.
Figure: Cloud Sequence Bio m5C Bis-Seq Experimental Procedure
3, cloud sequence biological RIP and RNA Pull down
When the sequencing, screening and verification work is over, if you want to publish high-scoring articles, then the functional mechanism experiments often have two good helpers: RIP and RNA pull down experiments, mainly to confirm that the candidate target genes are related to methylation. The direct binding of proteins is a powerful tool for resolving the regulation mechanism of RNA methylation. As in the above example, CancerCell, the authors demonstrated that the target gene regulated by RNA methylation can directly bind to the demethylase ALKBH5 through RIP and RNA pull down experiments.
So what is RIP and RNA pull down?
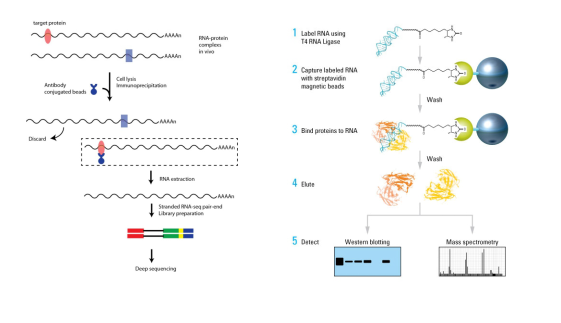
1RIP (RNA Immunoprecipitation) is an RNA immunoprecipitation technique that uses an antibody to specifically bind to a protein of interest to detect an RNA molecule that binds to the protein and is used to detect the interaction of a specific protein with RNA. RIP-Seq combines RIP technology with high-throughput sequencing to enable high-throughput detection of RNA bound to specific proteins, thereby resolving the target protein-RNA interaction network within the entire transcriptome. RIP-qPCR can detect RNA molecules bound to specific proteins. For example, whether a methylation modification-related enzyme binds to an RNA molecule can be detected by RIP-qPCR.
2RNA Pull down is the use of a specific probe to pull down the RNA of interest, and then detect the RNA or protein bound to it. Used to detect the interaction of specific RNAs with proteins/RNAs. RNA Pull down followed by Western Blot can detect whether a particular RNA interacts with a specific RNA-binding protein. RNA pull down followed by mass spectrometry experiments can resolve all protein information within the proteome range.
Figure: Cloud sequence biological RIP-seq and RNA Pull down experimental procedure
Sixth, RNA methylation project country natural application proposal
1. RNA methylation research is currently in the ascendant stage, and it is possible to try to design the research content of specific cell and tissue type expression profiles. Analyze the expression spectrum as early as possible, find the differentially modified molecules with high research value, enrich the functional prediction information, and carry out some simple verification experiments. At the same time, it is best to form a rough outline of functional mechanisms. It can also be tried in combination with other research hotspots, such as m6A modified miRNA expression profiles in exosomes.
2. Combine the modification enzymes of specific RNA methylation modification types to carry out functional mechanism research. In the direction of mechanism research, flexible use of a variety of research tools to effectively analyze the interaction of RNA molecules with other molecules or proteins. Including RIP, RNA pull-down and other experimental techniques, if you can get valid data, it will add a lot of color in the application of the National Natural Science Foundation.
Cloud order related product recommendation
m6A RNA methylation sequencing
m5C RNA methylation sequencing
RIP and RNA Pull down
Shanghai Yunxu Biological Technology Co., Ltd. Â
Shanghai Cloud-seq Biotech Co.,Ltd        Â
Address: 6/F, Building 71, Lane 1066, Qinzhou North Road, Caohejing High-tech Development Zone, Shanghai Â
Telephone website: Â
mailbox:
Next Article
What do you eat in the spring?